Kibble-Zurek quench in 2D transverse-field Ising model#
This example provides a quick overview of simulating the real-time Kibble-Zurek quench in 2D Ising model using YASTN library. We show an example of a system defined on a \(4{\times}4\) square lattice with open boundary conditions (OBC). The Hamiltonian reads
where \(\sigma^x\) and \(\sigma^z\) are standard Pauli matrices, and we assume random nearest-neighbor couplings \(J_{i,j} \in [-1, 1]\).
The amplitude of couplings is gradually turned on as \(f(s) = 1 + \sin(\pi (s - 0.5))\), and the transverse field is gradually turned off as \(g(s) = 1 - \sin(\pi (s - 0.5))\).
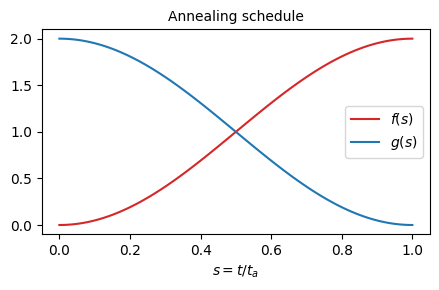
The system is initialized in the ground (product) state at \(s=0\), where the couplings \(f(0)=0\). The evolution ends upon reaching \(s=1\), where transverse field \(g(0)=0\). The quench rate is controlled by an annealing time \(t_a\) as \(s= t / t_a\). The system gets excited while passing the quantum critical point between paramagnetic and spin-glass phases in finite annealing time \(t_a\). This provides a minimal example of a class of problems considered in https://arxiv.org/abs/2403.00910
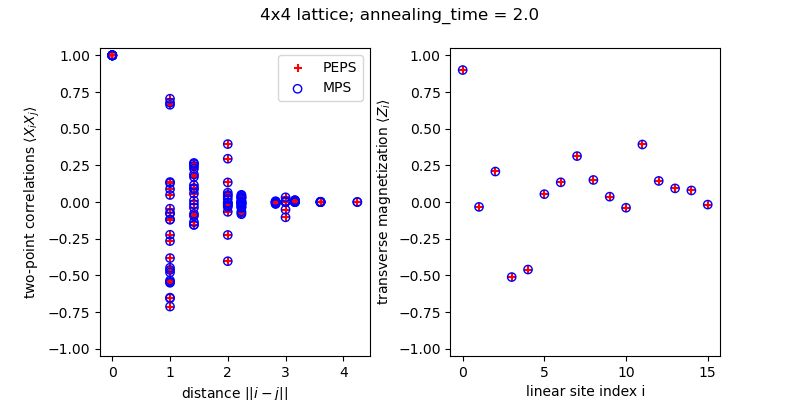
We compare the results obtained using MPS and PEPS routines. This example can be also run from tests/quickstart/test_KZ.py
- Initialization of Model Parameters:
import math import numpy as np import matplotlib.pyplot as plt from tqdm import tqdm # progressbar import yastn import yastn.tn.mps as mps import yastn.tn.fpeps as peps from yastn.tn.fpeps.gates import gate_nn_Ising, gate_local_field # # Employ PEPS lattice geometry for sites and nearest-neighbor bonds Nx, Ny = 4, 4 # lattice size geometry = peps.SquareLattice(dims=(Nx, Ny), boundary='obc') sites = geometry.sites() # list of lattice sites # # Draw random couplings from uniform distribution in [-1, 1]. np.random.seed(seed=0) Jij = {k: 2 * np.random.rand() - 1 for k in geometry.bonds()} # # Define quench protocol fXX = lambda s : 1 + math.sin((s - 0.5) * math.pi) fZ = lambda s : 1 - math.sin((s - 0.5) * math.pi) ta = 2.0 # annealing time dt = 0.04 # intended time step; make it smaller to decrease errors steps = round(ta / dt) # enforce integer number of Trotter steps dt = ta / steps # # Load operators. Problem has Z2 symmetry, which we impose. ops = yastn.operators.Spin12(sym='Z2')
- PEPS simulations; time evolution:
def gates_Ising(Jij, fXX, fZ, s, dt, sites, ops): """ Trotter gates at time s. """ gates = [] dt2 = 1j * dt / 2 for bond, J in Jij.items(): gt = gate_nn_Ising(J * fXX(s), dt2, ops.I(), ops.x(), bond) gates.append(gt) for site in sites: gt = gate_local_field(fZ(s), dt2, ops.I(), ops.z(), site) gates.append(gt) gates = gates + gates[::-1] # time-step is 1j * dt / 2, as we complete trotterized evolution # by its adjoint for 2nd order time-evolution method. return gates # # Initialize system in the product ground state at s=0. psi = peps.product_peps(geometry=geometry, vectors=ops.vec_z(val=1)) # # simulation parameters D = 6 # PEPS bond dimension opts_svd_ntu = {"D_total": D} # env = peps.EnvNTU(psi, which='NN+') # The environment used to calculate bond metric tensor. # This is a setup for NTU NN+ environment as described in # the appendix of https://arxiv.org/abs/2403.00910 # infoss = [] # for diagnostics information # # execute time evolution t = 0 for _ in tqdm(range(steps)): t += dt / 2 gates = gates_Ising(Jij, fXX, fZ, t / ta, dt, sites, ops) infos = peps.evolution_step_(env, gates, opts_svd=opts_svd_ntu) # The state psi is contained in env # evolution_step_ updates psi in place. infoss.append(infos) t += dt / 2 Delta = peps.accumulated_truncation_error(infoss, statistics='mean') print(f"Accumulated mean truncation error: {Delta:0.5f}")
- PEPS simulations; final correlations:
# We employ boundary MPS to contract the network opts_svd_env = {'D_total': 4 * D} opts_var_env = {"max_sweeps": 8, "overlap_tol": 1e-5, "Schmidt_tol": 1e-5} # # setting-up environment env_mps = peps.EnvBoundaryMPS(psi, opts_svd=opts_svd_env, opts_var=opts_var_env, setup='lr') # # Calculating 1-site <Z_i> for all sites Ez_peps = env_mps.measure_1site(ops.z()) # # Calculating 2-site <X_i X_j> for all pairs i <= j Exx_peps = env_mps.measure_2site(ops.x(), ops.x(), opts_svd=opts_svd_env, opts_var=opts_var_env)
- MPS simulations:
# Map between sites and linear MPS ordering. s2i = {s: i for i, s in enumerate(sites)} # # Map for bonds, sorting pairs of MPS indices for convinience b2i = lambda s1, s2: tuple(sorted([s2i[s1], s2i[s2]])) # # define Hamiltonian MPO HI = mps.product_mpo(ops.I(), N=Nx*Ny) # identity MPO # termsXX = [mps.Hterm(amplitude=J, positions=[s2i[s1], s2i[s2]], operators=[ops.x(), ops.x()]) \ for (s1, s2), J in Jij.items()] HXX = mps.generate_mpo(HI, termsXX) # termsZ = [mps.Hterm(-1, i, ops.z()) for i in range(Nx * Ny)] HZ = mps.generate_mpo(HI, termsZ) # # MPO contributions in H(t) will be added up. H = lambda t: [HXX * fXX(t / ta), HZ * fZ(t / ta)] # # Initial state. TDVP is unstable starting in a product state # There are many strategies to mitigate it. # Here, a simple strategy to start with a product state obtained # via DMRG with artificially enlarged bond dimension is sufficient. psi = mps.random_mps(HI, D_total=16) # initialize with D=16 mps.dmrg_(psi, H(0), method='1site', max_sweeps=8, Schmidt_tol=1e-8) # # time-evolution generator and its parameters opts_expmv = {'hermitian': True, 'tol': 1e-12} opts_svd = {'tol': 1e-6, 'D_total': 64} # max MPS bond dimension evol = mps.tdvp_(psi, H, times=(0, ta), method='12site', dt=dt, order='2nd', opts_svd=opts_svd, opts_expmv=opts_expmv, progressbar=True) # # run evolution # evol is a generator with one (final) snapshot to reach next(evol) # execute time evolution # # calculate expectation values Ez_mps = mps.measure_1site(psi, ops.z(), psi) Exx_mps = mps.measure_2site(psi, ops.x(), ops.x(), psi, bonds="<=")
- Compare results of PEPS and MPS:
Z_peps = np.array([Ez_peps[st].real for st in sites]) Z_mps = np.array([Ez_mps[s2i[st]].real for st in sites]) error_Z = np.linalg.norm(Z_peps - Z_mps) / np.linalg.norm(Z_mps) print(f"Relative difference of PEPS vs MPS in Z magnetization: {error_Z:0.5f}") # Euclidian distance on a square lattice dist = lambda s1, s2: np.linalg.norm([s1[0]-s2[0], s1[1]-s2[1]]) rs = np.array([dist(s1, s2) for (s1, s2) in Exx_peps]) # XX_peps = np.array([*Exx_peps.values()]).real XX_mps = np.array([Exx_mps[b2i(*bond)] for bond in Exx_peps.keys()]).real error_XX = np.linalg.norm(XX_peps - XX_mps) / np.linalg.norm(XX_mps) print(f"Relative difference of PEPS vs MPS in XX correlations: {error_XX:0.5f}")
- Visualize:
fig, ax = plt.subplots(1, 2) fig.set_size_inches(8, 4) plt.subplots_adjust(hspace=0.3, wspace=0.3) ax[0].scatter(rs, XX_peps, label='PEPS', marker='+', color='r') ax[0].scatter(rs, XX_mps, label='MPS', marker='o', color='b', facecolors='none') ax[0].set_ylim([-1.05, 1.05]) ax[0].set_xlabel(r"distance $||i - j||$") ax[0].set_ylabel(r"two-point correlations $\langle X_i X_j \rangle$") ax[0].legend() ax[1].scatter(np.arange(len(Z_peps)), Z_peps, label='PEPS', marker='+', color='r') ax[1].scatter(np.arange(len(Z_mps)), Z_mps, label='MPS', marker='o', color='b', facecolors='none') ax[1].set_xlabel(r"linear site index i") ax[1].set_ylabel(r"transverse magnetization $\langle Z_i \rangle$") ax[1].set_ylim([-1.05, 1.05]) fig.suptitle(f"{Nx}x{Ny} lattice; annealing_time = {ta:0.1f}") fig.show()